Introduction

Preserving animal and plant diversity is an increasingly important task in the face of the massive destruction and alteration of natural landscapes. In this perspective, attention is being focused upon rare and endangered insect species, particularly butterflies. But conservation should act at both species and gene diversity level. Polymorphism is generally thought to improve fitness and ability to face environmental changes and it is predicted that the reduction of population size and interdemic gene exchange will in time cause progressive impoverishment in a gene pool resulting in inbreeding (Kaidanov, 1995).
Black musk swallowtail (Atrophaneura alcinous Klug.) - is a unique representative of Troidini at the Russian Far East (Korshunov and Gorbunov, 1994). The tribe includes the most large and attractive papilionid species and many of them are rare or endangered and are listed in Red Data Books and CITES.
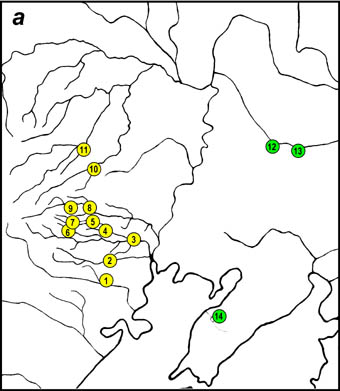
At the Russian Far East black musk swallowtail is presented by subspecies A. a. confusus, which is treated as rare, and is in need of protection and listed in Russian Red Data Book. Sporadic distribution of musk swallowtail is due to the dot distribution pattern of its hostplant Aristolochia manshuriensis, which mainly grows in the area of Shufan Plateau (valleys of the rivers Borisovka (Shufan), Krounovka (Chapigou), Nezhinka (Sanduga), Anan'evka (Elduga), Gryaznaya, Amba. A. manshuriensis also grows in implantations, where A. alcinous gives temporary populations (Botanical Garden in Vladivostok's suburb, De-Friz Peninsula, Gornotayozhnoe and Kamenushka, district of Ussuriisk).
The objective of this study was to estimate RAPD-polymorphism and morphological variability in natural habitats of black musk swallowtail and in the colonies appeared in artificial implantations of the hostplant.
Materials and methods
Entomological material was collected with conventional techniques in Primorye in 1998-1999. Specimens of A. a. alcinous were kindly provided by Shigeru Albert Ae (Nagoya city, Japan). Adults were caught with a butterfly net. Eggs and larvae were also collected and reared to adults in captivity.
Total genomic DNA was extracted by standard phenol-detergent technique. Samples were incubated with proteinase K. We did not treated samples with RNAse.
Amplification reactions were carried out according to the main recommendations given by Williams et al. (1991). Reaction mixture in the volume of 25 μl contained 10 mM Tris-HCl, pH 9.0; 50 mM KCl; 1.9 mM MgCl2; 0.01 % gelatin; 0.1 % Triton X-100; 125 μM each dNTP; 0.25 μM primer ("Operon Technologies", Alameda, USA); 0.5 unit of Taq DNA polymerase; and 10-15 ng genomic DNA. Amplifications were performed in UNO Termoblock thermal cycler (Biometra, Germany ) for 1 minute at 94°C (initial denaturation); 45 cycles of 1 minute at 94°C; 1 minute at 35°C (primer annealing step); 2 minutes at 72°C (elongation step); 5 minutes at 72°C (final elongation step).
Amplification products were analyzed by electrophoresis in 1.8 % agarose gels in 1´ TBE and detected by ethidium bromide staining. Further analysis of the data was performed using TFPGA (Miller, 1997) and POPGENE (Yeh et al., 1999) computer software.
Estimation of morphological variability was carried out by discriminant analysis of 22 biometrical wing pattern characters with the use of Statistica for Windows 5.0.
Results
Fifty 10-base primers were initially screened for their ability to prime PCR amplification of A. alcinous DNA. Only 33 RAPD primers (66%) yielded amplification products, while the rest of the primers did not amplify the DNA template or resulted in smear or faint bands. Five positive primers that gave reproducible RAPD patterns with resolvable fragments were selected for further analysis.
Amplification products ranged in size from 200 to 2500 bp. The percentage of polymorphic bands was about 50 %. Comparison of RAPD-patterns revealed specific differences between A. a. alcinous and A. a. confusus : fragments OPE-041100 and OPE-17370 were consistently amplified in nominative subspecies (Fig. 1). Most bands appeared in the individuals with high frequency. Markers for local colonies of A. a. confusus were not detected.
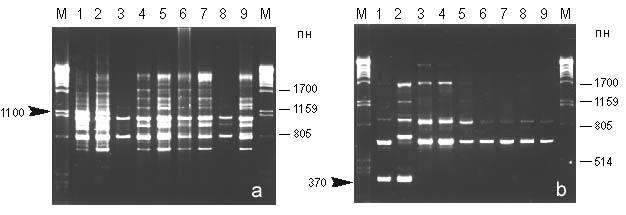
Fig. 1. RAPD-patterns amplified with primers OPE-04 (a) and OPE-17 (b) of Atrophaneura alcinous . Lane M, Lambda phage DNA digested with PstI; lanes 1-2, A. a. alcinous; 3-9, A. a. confusus. Arrows indicate specific markers.
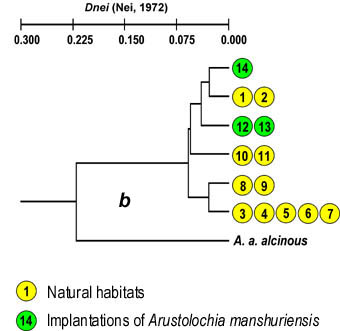
The values of Nei's genetic distances (1972) were very small for local colonies in Primorye and ranged from 0.024 to 0.072, while genetic distances for A. a. confusus and nominative subspecies A. a. alcinous were from two to ten times higher and ranged from 0.153 to 0.259. UPGMA dendrogram based on Nei's genetic distances demonstrates similarity of subpopulations (Fig. 2).