Introduction
 Parnassius bremeri and P. nomion, like
other representatives of Parnassiini tribe (family Papilionidae), show
extremely high level of individual and geographic variation. Distribution
of these species covers Zabaikalye, Priamurye, Primorye, Kunashir Island
(P. bremeri only), North-East China, North
Korea and Japan (Hokkaido). Areas of two species overlap essentially in
this territory that with weak mechanisms of reproductive isolation favors
hybridization between these species in nature.
Parnassius bremeri and P. nomion, like
other representatives of Parnassiini tribe (family Papilionidae), show
extremely high level of individual and geographic variation. Distribution
of these species covers Zabaikalye, Priamurye, Primorye, Kunashir Island
(P. bremeri only), North-East China, North
Korea and Japan (Hokkaido). Areas of two species overlap essentially in
this territory that with weak mechanisms of reproductive isolation favors
hybridization between these species in nature.
Existence of presumed hybrids between P. nomion and P. bremeri was described as long ago as the beginning of the century (Verity, 1907; Bryk, 1935). Recently identification of hybrids between P. nomion and P. bremeri was maid by discriminant analysis (Glushchenko, Martynenko, 1997).
Our objectives were to carry out morphological identification of hybrid specimens and to reveal genetic evidences of hybridization.
Materials and methods
Entomological material collected in the vicinities of Sadovoe (District of Dal'negorsk, Primorye) and Shumny (District of Chuguevka) was kindly provided by Yu.N. Glushchenko and A.B. Martynenko. Ten specimens of P. bremeri and fourteen specimens of P. nomion were sampled in four different locations. Standard discriminant function analysis of 27 morphometric characters was used to identify phenotypical hybrids.
A total of twenty-eight specimens were used for RAPD analysis. Each specimen was given a numerical identification for cross-referencing with molecular data. The thorax of each butterfly was used for DNA extraction, and the abdomen, head, wings and legs were saved as vouchers.
Total genomic DNA was extracted from thoraces of individual adult butterflies by standard phenol-detergent technique.
RAPD amplification was mainly done according to Williams et al. (1991). Polymerase chain reaction conditions of the RAPD reactions were optimized to yield reproducible results.
The usual precautions needed to prevent contamination of test specimens with foreign DNA and previously amplified fragments included physical separation of specimen dissection, DNA extraction and amplification areas, sterile technique by using laminar flow hood, and use of sterile disposable tubes and tips. To test the reality of PCR products the control with no genomic DNA was routinely used.
Amplification products were analyzed by electrophoresis in 2 % agarose gels and detected by staining with ethidium bromide. Sizes of amplified fragments were estimated by comparison with lambda DNA digested with PstI or EcoRI and HindIII.
Results and discussion
The discriminant model was calculated on the base of standard discriminant analysis. Four specimens identified as hybrids originated from the vicinity of Sadovoe. Confident differences between parent species and hybrids were observed (Fig. 1).
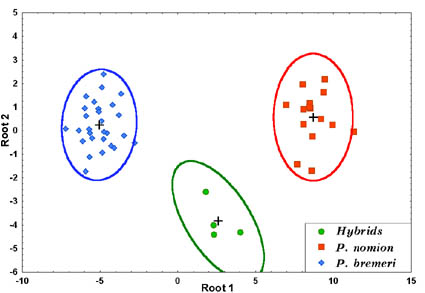
Fig. 1. Distribution of studied specimens in the two-dimensional scale of canonical variables.
To reveal genetic evidences of hybridization RAPD-PCR and PCR-RFLP were used. A total of 25 primers obtained from Operon Technologies Inc. (Alameda, CA) were screened against two specimens of P. nomion and P. bremeri. Among these primers sixteen were initially appeared to amplify diagnostic markers and were tested against larger selection of specimens collected in distinct isolated populations of each species.
Ten primers were selected because of its efficiency in the characterization of samples. They were tested both on specimens collected from distinct populations (Luk'yanovka, Shumny, middle stream of the River Krasnaya) and specimens from the vicinity of Sadovoe where hybridization occurred.
Comparison of amplification results has shown that both P. bremeri and P. nomion exhibit presence of high variation among individuals within the species. In P. nomion most amplified fragments were shared among individuals with high frequency but they were not species specific. Some PCR products were unique to individuals, and some were species-specific. In P. bremeri most PCR-products were unique to individuals, but there were also DNA fragments recorded in all individuals of this species. Presence of such conserved fragments allowed using them as potential species-specific markers.
Only three primers (OPA-08, OPA02, OPB-08) screened against 15 specimens of two species collected in isolated populations initiated synthesis of diagnostic species-specific markers. Primer OPA-08 consistently amplified an approximately 500 basepair fragment in P. nomion and an approximately 400 basepair fragment in P. bremeri (Fig. 2). Primer OPA-02 initiated amplification of an approximately 300 basepair fragment in P. nomion. Primer OPB-08 consistently amplified an approximately 2000 fragment in P. nomion and an approximately 1300 basepair fragment in P. bremeri. These fragments (OPA-08400, OPA-08500, OPA-02300,OPB-081300, OPB-082000) named according to the convention suggested by Paran et al. (1991) were considered as species-specific markers and were used for identification of phenotypically presumed hybrids.
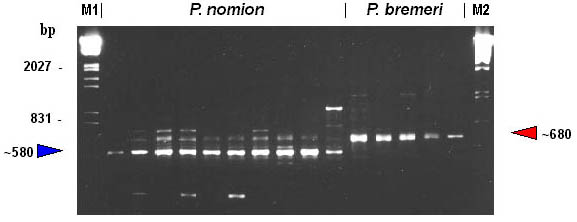
Fig. 2. RAPD-patterns amplified with primer OPA-08. M1, EcoRI+HindIII digests of Lambda phage DNA. M2, PstI digests of Lambda phage DNA.
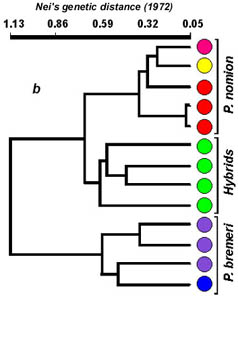
By the features of RAPD pattern, phenotypical hybrids take up an intermediate position between P. nomion and P. bremeri as they show whether a complete additive pattern of species specific markers or presence of different diagnostic fragments that are species-specific in high frequency. Complete additive patterns were observed only with primer OPA-08 (Fig. 3). Other two primers (OPA-02, OPB-08) gave different diagnostic fragments shared among hybrid specimens and parent species that also supports hybrid origin of tested specimens.
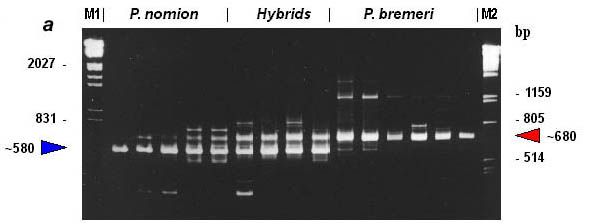
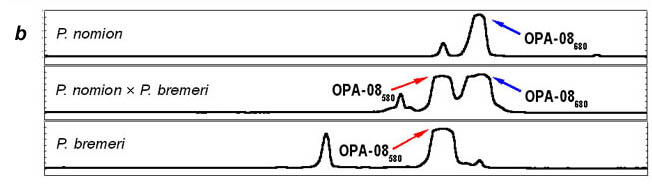
Fig. 3. RAPD-evidence of hybridization between Parnassius nomion and P. bremeri : electrophoregram (a) and profiles of specific patterns (b). M1, EcoRI+HindIII digests of Lambda phage DNA. M2, PstI digests of Lambda phage DNA.
Band-sharing analysis was performed on the results of RAPD-patterns obtained using five primers. All gel patterns were visually aligned to establish homologous bands, and RAPD patterns were compared. A total of 195 RAPD bands were scored, including all but very weak ones. Genetic distances (Nei, 1972) were calculated using RAPDist program ver. 1.04.
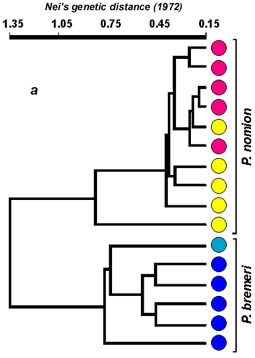
Basing on calculated distance matrix, average inter- and intraspecific distances were calculated for both species. Average distance within P. bremeri is considerably higher in comparison with P. nomion, and equals 0.537, against 0.374 in P. nomion. Average interspecific distance for P. nomion and P. bremeri is 1.352. Average value of genetic distance between hybrid individuals is 0.552, meanwhile distances between hybrids and P. nomion and between hybrids and P. bremeri are 0.658 and 0.838 accordingly. It means that hybrid specimens are more close to P. nomion, than to P. bremeri. Nevertheless, these distance values are within the range of interspecific differences. Extremely high level of genetic polymorphism revealed in P. bremeri with average value of intersubspecific genetic distances similar to that of hybrid specimens (0.537 and 0.552) may be treated as corroboration of hybrid origin of the subspecies P. b. orotschonicus, that was assumed by Kurentsov (1974).
Cluster analysis of distance measures via UPGMA (Unweighted Pair Group Method of Arithmetic means) was carried out using a computer program NTSYS-pc (Fig. 4).
|
|
|
Cluster analysis revealed that all tested specimens of both Parnassius species are separated in two well-differentiated groups. Cluster of hybrid specimens is well differentiated from P. bremeri but is more close to a second parent species P. nomion that may be explained with some ecological traits of hybridization. Flight periods of these two species overlap in the area of hybridization. Females of P. bremeri continue to emerge from pupae even after all males of this species have died. In this time males of P. nomion begin to emerge and they become exposed to a deficit of females for mating. It makes favorable conditions for interspecific mating with females of P. bremeri. Almost all specimens tested by RAPD analysis were males and we could not reveal any sex-linked markers. Nevertheless, we consider that closer relation of hybrid specimens to one parent species P. nomion is explained by unidirectional hybridization between two species. This presumption is also supported by the data of PCR-RFLP (2.3 kb COI & COII mtDNA genes) as all hybrids have P. bremeri haplotypes. No evidences of introgression were revealed on the material studied. Nevertheless, the possibility of introgression remains and more data must be obtained to reveal it.
Being based on the results received, we can assert that the specimens with atypical for pure forms set of morphological characters are really hybrids between two species P. bremeri and P. nomion. It proves the fact of their natural hybridization in the location where samples were collected. Basing on previously published notes and some personal communications provided by other researchers about findings of similar hybrid-like specimens in other parts of the area of these two species, we can presume that natural hybridization is a quite common phenomena in some localities where natural conditions are favorable for hybridization.
References
Bryk F. (1935) Lepidoptera. Parnassiidae, pars 2 (Subfamilia Parnassiinae). Das Tierreich. 65 Lieferung. Berlin, Leipzig. 790 p.k
Glushchenko Yu.N., Martynenko A.B. (1997) Hybrids between Parnassius bremeri and Parnassius nomion in collections from southern Far East and Zabaikalye. Animals and plants of the Far East. Vol. III. P. 31-37. [In Russian]
Kurentsov A.I. (1970) The butterflies of the Far East USSR (Key). Leningrad, Nauka. 163 p. [In Russian]
Verity R. (1905-1911) Rhopalocera palearctica: Iconographie et description des papilons diurnes de la region palearcticque. Papilionidae et Pieridae. Florence: Verity; Landi. 386 p.
Williams J.G.K., Kubelik A.R., Livak K.J., Rafalski J.A., and Tingey S.V. (1991) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research, 18: 6531-6535.